Computational Modeling
NREL uses computational modeling to increase the efficiency of biomass conversion by rational design using multiscale modeling, applying theoretical approaches, and testing scientific hypotheses.
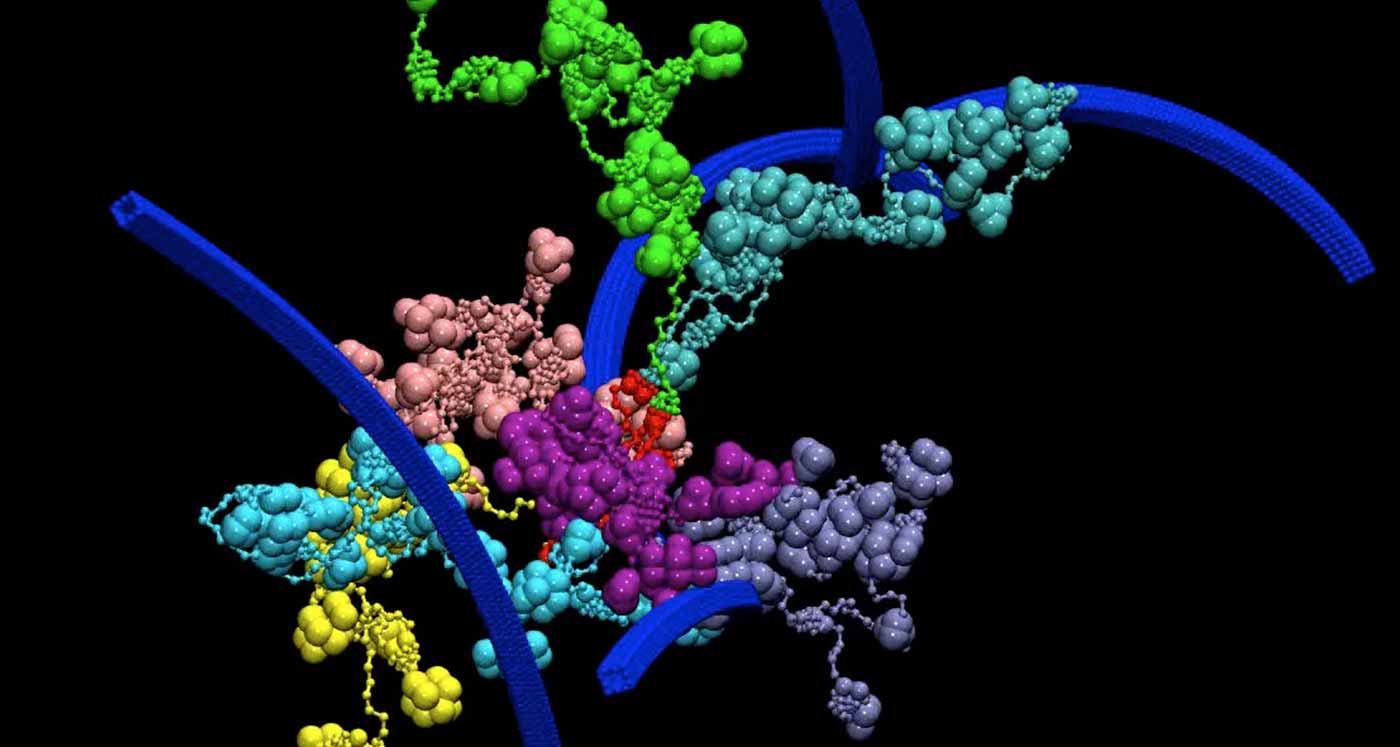
Cellulosomes are complexes of protein scaffolds and enzymes that are highly effective in decomposing biomass. This is a snapshot of a coarse-grain model of complex cellulosome binding to a coarse grain model of cellulose fibrils. The complex cellulosome is constructed of seven cellulosomes (green, blue, cyan, pink, purple, majenta, and yellow) bound to a secondary scaffold (red) by cohesin-dockerin pairs. The secondary scaffold is very flexible and allows for motion of the primary cellulosomes within a range. The primary cellulosomes are made of nine enzymes each bound to a primary scaffold via cohesin-dockerin pairing with many flexible linkers. All primary scaffolds and some enzymes have cellulose binding domains for attaching to cellulose.
Featured Publications

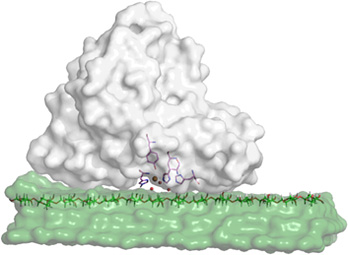
Predicting Enzyme Adsorption to Lignin Films by Calculating Enzyme Surface Hydrophobicity, The Journal of Biological Chemistry (2014)
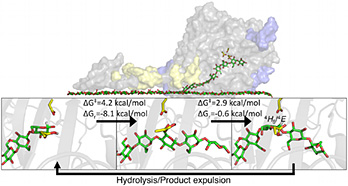
CarbohydrateProtein Interactions That Drive Processive Polysaccharide Translocation in Enzymes Revealed from a Computational Study of Cellobiohydrolase Processivity, Journal of the American Chemical Society (2014)
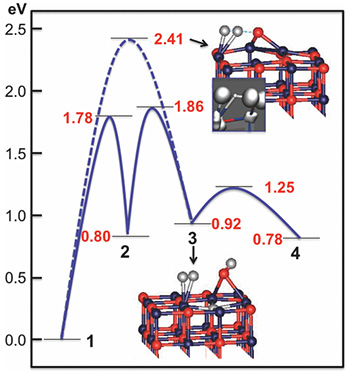
Initial reduction of the NiO(100) surface in hydrogen, The Journal of Chemical Physics (2013)
View all NREL bioenergy computational modeling publications.
Capabilities
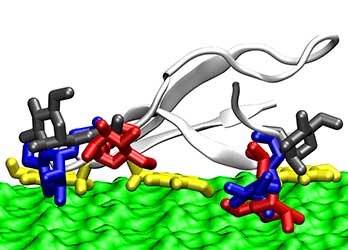
Computer Enzyme Design
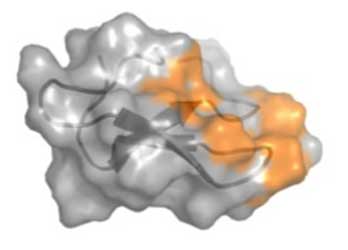
NREL's molecular modeling provides insight into enzyme mechanisms and enables higher performance and new functionality for industrial enzymes. For example, the study depicted by the illustration to the left determined the structure and binding orientation of several glycosylation patterns on the binding domain, which we showed in previous studies to greatly enhance the binding to cellulose, a property that has been shown to improve the enzyme activity in breaking down cellulose in biomass.
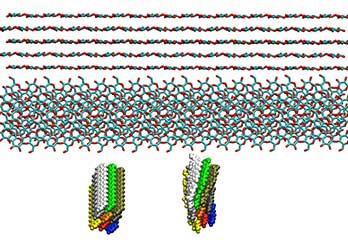
Cell Wall Components
We use multi-scale modeling to investigate the properties of cellulose, hemicellulose, lignin, and pectin. Together they compose cell walls and are the source of biofuels and biomaterials. Our modeling investigates their properties for better conversion and material design.
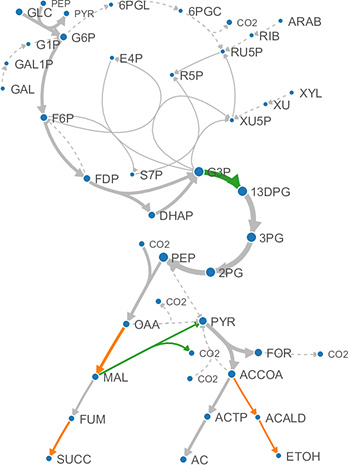
Metabolic Modeling
To increase yields and redirect production to desired products, we model the existing pathways of microbial conversion of sugars to fuels and high-value products.
Quantum Mechanical Models
NREL studies chemical and electronic properties and processes to reduce barriers in conversion and upgrading of renewable sources.
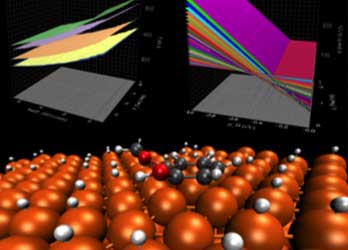
Catalyst Science and Design
We use ab initio reaction energetics coupled with experimental kinetic models to predict catalyst performance under realistic reaction conditions.
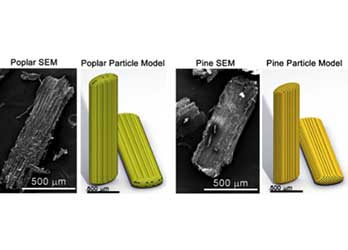
Biomass Mesostructure and Continuum Multiphysics Modeling
We are developing realistic, species-specific particle models that predict how feedstocks and blendstocks will perform in various conversion processes.
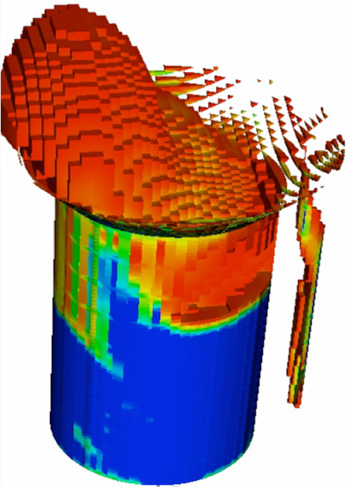
Reactor Simulations
We use multiphase (solid, liquid, and/or s + gas) flow simulations to describe, understand, and optimize reactors used in catalytic pyrolysis, enzymatic hydrolysis, and fermentation. Efforts also include coupling flow models with reaction kinetics and interphase transport. experiments.
Research Team
Principal Investigators
Peter Ciesielski
Research Scientist, Bioenergy and Biomaterials
Peter.Ciesielski@nrel.gov303-384-7691
Lintao Bu
James Lischeske
Ambarish Nag
Hariswaran Sitaraman
Mike Sprague
Related and Integrated Programs
Biological and Catalytic Conversion of Sugars and Lignin
Biomass Deconstruction and Pretreatment
Enzyme and Microbial Development
Heterogeneous Catalysis for Thermochemical Conversion
Thermochemical Process Integration, Scale-Up, and Piloting
Collaborators
BioEnergy Science Center
C3Bio: Center for Direct Catalytic Conversion of Biomass to Biofuels
Computational Pyrolysis Consortium
Share