
Research Interests
-
Development of thermophilic bacteria for improved biomass deconstruction
-
Biomass degrading mechanisms of thermophilic bacteria
-
Metabolic modeling and pathway engineering in microbes for upgrading sugar to fuels and high value chemicals
-
Metabolic enzyme characterization, modeling, and engineering (thermostability, cofactor specificity, binding, rate limiting step)
-
Characterization, modeling, and engineering of biomass degrading enzymes from mesophilic and thermophilic bacteria
-
Quantum chemical calculations for the study of thermochemical properties and processes
Affiliated Research Programs
-
Center for Bioenergy Innovation
-
Metabolic Modeling and Pathway Engineering
-
Mechanism and Enhancement of Process-Relevant Enzymes
-
Enzyme Engineering and Optimization
-
Targeted Microbial Development
-
Lignin Utilization
Areas of Expertise
-
Molecular dynamics
-
Quantum chemistry
-
Computational modeling
-
Computer software development (for applications in molecular dynamics [MD] and quantum mechanics [QM] calculations)
-
Protein purification
-
Protein characterization
-
Metabolic modeling
Education
-
Ph.D., Chemical Physics, University of Texas, Austin, 2006
-
IMCC exchange fellow, Université de Lille I/ University of Texas, Austin, 2001
-
B.S., Physics (with honors), minors in Mathematics and Chemistry, Université de Lille I, Lille, France, 2001
-
Mathématiques Speciales, Lycée Faidherbe, Lille, France, 1999
-
Mathématiques Superieures, Lycée Faidherbe, Lille, France, 1998
Professional Experience
-
Senior Research Scientist, National Renewable Energy Laboratory (NREL), 2010–present
-
Research Scientist, NREL, 2008–2010
-
Research Associate, Scripps Research Institute, Department of Molecular Biology, 2006–2008
-
Graduate Research Assistant, University of Texas, Department of Chemistry and Biochemistry, 2001–2006
-
Visiting Research Fellow, Universitt Mainz, Germany, Institut für Physikalische Chemie, 2004 and 2005
Featured Publications
-
"The Multi Domain Caldicellulosiruptor bescii CelA Cellulase Excels at the Hydrolysis of Crystalline Cellulose," Scientific Reports (2017)
-
"A quantitative model for the prediction of sooting tendency from molecular structure," Energy & Fuels (2017)
-
"Structural, Mutagenic and In Silico studies of Xyloglucan Fucosylation in Arabidopsis thaliana Suggest a Water‐Mediated Mechanism," The Plant Journal (2017)

-
"In vivo Synergistic Activity of a CAZyme Cassette from Acidothermus cellulolyticus Significantly Improves the Cellulolytic Activity of the C. bescii Exoproteome," Biotechnology and Bioengineering (2017)
-
"Metabolic engineering of Actinobacillus succinogenes provides insights into succinic acid biosynthesis," Applied and Environmental Microbiology (2017)
-
"Engineering the N‐terminal end of CelA results in improved performance and growth of Caldicellulosiruptor bescii on crystalline cellulose," Biotechnology and Bioengineering (2017)
-
"Multifunctional Cellulolytic Enzymes Outperform Processive Fungal Cellulases for Coproduction of Nanocellulose and Biofuels," ACS nano (2017)
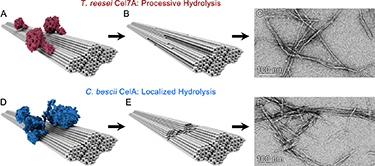
-
"Efficient estimation of the maximum metabolic productivity of batch systems," Biotechnology for Biofuels (2017)
![Flow chart using six graphics that show how productivity–yield surfaces are calculated. The upper left figure labeled (a) Metabolic Model shows two green dots on the left with arrows pointing into a circle with four blue dots on the circumference of a circle with arrows cycling clockwise and one blud dot in the center of the circle that is also involved in the clockwise cycle. Two of the outer blue dots point left to three other blue dots that point outside of the larger circle to three green dots. This image points to the upper and lower center images. The upper center image is labeled (c) EFM Selection and shows a graph with Biomass Formation as the y-axis and Product Formation as the x-axis with a scattering of blue dots on a blue background with seven red dots on the periphery of the blue background. This upper center image points to the upper right image, which is labeled (e) Productivity Maximum and shows a graph with [S] as the y-axis and Time as the x-axis. The graph is separated into three blue rectangular areas and two yellow rectangular areas with five series of three green dots connected by a green line going up from left to right and five series of three red dots connected by a red line going down from left to right. This image points down to the lower right image. The lower left image is labeled (b) Growth Rate Inhibition and shows a graph with a y-axis labeled micron and an x-axis labeled [Substrate] with three lines (the top is green the middle is blue, and the bottom is red) connecting dots going down from left to right. This image points to the lower center image. The lower center image is labeled (d) Uptake Dynamic Model and shows a graph with [S] as the y-axis and Time as the x-axis with two lines connecting dots, the red line goes down from left to right and the green line goes up from left to right. This image points to the upper right image. The lower right image is labeled (f) Productivity-Yield Surface and is a graph with a y-axis labeled Yield and an x-axis labeled Productivity with three lines (the top is red, the middle is green, and the bottom is blue) connecting dots going down from left to right, with grey vertical lines coming down from the blue connected dots.](/bioenergy/assets/images/bomble_estimation_max_metabolic_productivity_pub1.jpg)
-
"Heterologous Expression of Two Different Family 10 Xylanases from Acidothermus cellulolyticus Enhances the Exoproteome of Caldicellulosiruptor bescii and Growth on Xylan Substrates," Biotechnology for Biofuels (2016)
-
"Comparing Residue Clusters from Thermophilic and Mesophilic Enzymes Reveals Adaptive Mechanisms," PloS One (2016)
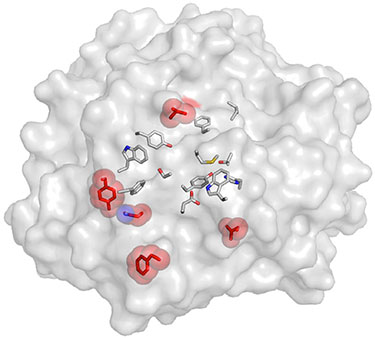
-
"Strategies to reduce end-product inhibition in family 48 glycoside hydrolases," Proteins (2016)
-
"Dramatic performance of Clostridium thermocellum explained by its wide range of cellulase modalities," Science Advances (2016)
-
"Chapter 7: New Insights into Microbial Strategies for Biomass Conversion," in Direct Microbial Conversion of Biomass to Advanced Biofuels (2015)
-
"Chapter 15: Influence of Particle Size on Direct Microbial Conversion of Hot-Water Pretreated Poplar by Clostridium thermocellum," in Direct Microbial Conversion of Biomass to Advanced Biofuels (2015)
-
"New perspective on glycoside hydrolase binding to lignin from pretreated corn stover," Biotechnology for Biofuels (2015)
-
"Expression of the Acidothermus cellulolyticus E1 Endoglucanase in Caldicellulosiruptor bescii Enhances its Ability to Deconstruct Crystalline Cellulose," Biotechnology for Biofuels (2015)
-
"The catalytic mechanism and unique low pH optimum of Caldicellulosiruptor bescii family 3 pectate lyase," Acta Crystallographica Section D (2015)
-
"Cofactor Specificity of the Bifunctional Alcohol and Aldehyde Dehydrogenase (AdhE) in Wild-Type and Mutant Clostridium thermocellum and Thermoanaerobacterium saccharolyticum," Journal of Bacteriology (2015)
-
"Homologous expression of the Caldicellulosiruptor bescii CelA reveals that the extracellular protein is glycosylated," PloS One (2015)
-
"Deletion of Caldicellulosiruptor bescii CelA reveals its crucial role in the deconstruction of lignocellulosic biomass," Biotechnology for Biofuels (2014)
-
"Predicting Enzyme Adsorption to Lignin Films by Calculating Enzyme Surface Hydrophobicity," Journal of Biological Chemistry (2014)
-
"Response to Comment on “Revealing Nature’s Cellulase Diversity: The Digestion Mechanism of Caldicellulosiruptor bescii CelA," Science (2014)
-
"Experimental and modeling studies of an unusual water-filled pore structure with possible mechanistic implications in family 48 cellulases," Journal of Physical Chemistry B (2014)
-
"Chapter 2: Self Assembly and Application of Cellulosomal Components," in Bionanotechnology: Biological Self-assembly and its Applications (2013)
-
"Revealing Nature's Cellulase Diversity: The Digestion Mechanism of Caldicellulosiruptor bescii CelA," Science (2013)
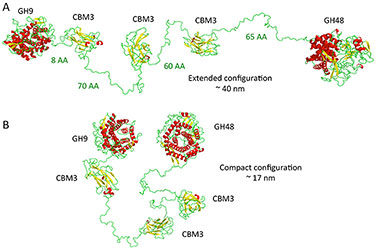
-
"Improving activity of minicellulosomes by integration of intra- and intermolecular synergies," Biotechnology for Biofuels (2013)
-
"Molecular dynamics simulations of the interaction of glucose with imidazole in aqueous solution," Carbohydrate Research (2012)
-
"Structure and function of the Clostridium thermocellum cellobiohydrolase A X1-module repeat: enhancement through stabilization of the CbhA complex," Acta Crystallographica Section D (2012)
-
"Computational study of bond dissociation enthalpies for a large range of native and modified lignins," Journal of Physical Chemistry Letters (2011)
-
"Multiple functions of aromatic-carbohydrate interactions in a processive cellulase examined with molecular simulation," Journal of Biological Chemistry (2011)
-
"Molecular-level origins of biomass recalcitrance: decrystallization free energies for four common cellulose polymorphs," Journal of Physical Chemistry B (2011)
-
"Applications of computational science for understanding enzymatic deconstruction of cellulose," Current Opinion in Biotechnology (2011)
-
"Chapter 4: Modeling the Cellulosome Using Multiscale Methods," in Computational Modeling in Lignocellulosic Biofuel Production (2010)
-
"Modeling the Self-assembly of the Cellulosome Enzyme Complex," Journal of Biological Chemistry (2010)
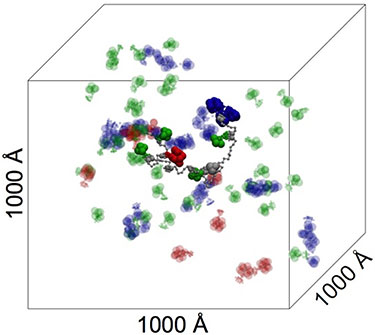
-
"The O-Glycosylated Linker from the Trichoderma reesei Family 7 Cellulase Is a Flexible, Disordered Protein," Biophysical Journal (2010)
-
"The unique binding mode of the Cellulosomal CBM4 from Clostridium thermocellum Cellobiohydrolase A," Journal of Molecular Biology (2010)
-
"Identification of Amino Acids Responsible for Processivity in a Family 1 Carbohydrate-Binding Module from a Fungal Cellulase," Journal of Physical Chemistry B (2010)
-
"The energy landscape for the interaction of the family 1 carbohydrate-binding module and the cellulose surface is altered by hydrolyzed glycosidic bonds," Journal of Physical Chemistry B (2009)
-
"Multiscale model of nucleic acids: insights into DNA flexibility," Biopolymers (2008)
-
"Factors Contributing to the Accuracy of Harmonic Force Field Calculations," Journal of Chemical Theory and Computation (2007)
-
"Thermochemistry of Key Soot Formation Intermediates: Isomers of C3H3," Journal of Physical Chemistry A (2007)
-
"High-Accuracy Extrapolated Ab Initio Thermochemistry. II: Minor Improvements to the Protocol and a Vital Simplification," Journal of Chemical Physics (2006)
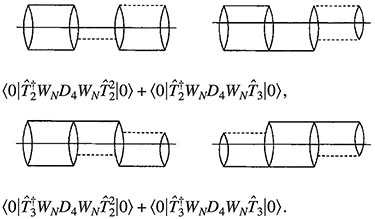
-
"Computer Software Review: ADF 2005 (Amsterdam Density Functional 2005)," Journal of American Chemical Society (2006)
-
"Coupled-cluster methods including non iterative Approximate quadruple excitation corrections," Journal of Chemical Physics (2005)
-
"Equation-of-motion coupled-cluster methods for ionized states with an approximate treatment of triple excitations," Journal of Chemical Physics (2005)
-
"Self-assembling dimeric and trimeric aggregates based on solvophobic and charge-pairing interactions," Supramolecular Chemistry (2004)
-
"On the vertical excitation energy of cyclopentadiene," Journal of Chemical Physics (2004)
- "Threshold detection using indicator-displacement assays: An application in the analysis of malate in Pinot Noir grapes," Journal of American Chemical Society (2004)
View all NREL Publications for Yannick J. Bomble.
Please contact me with research questions or ideas for collaborations. Please do NOT contact me directly for jobs—see instead information on NREL's Director's Postdoctoral Fellowship program or on NREL Careers in general.
Share
