
Research Interests
David Mulder's research interests revolve around the mechanisms of redox enzymes that catalyze primary oxidation-reduction reactions for solar energy conversion and biofuel production. Of particular interest is to understand the physical properties of biological redox cofactors and the underlying mechanistic principles that govern electron-transfer reactions and the transformation of electrochemical and photochemical energy into chemical bonds. David has technical expertise in applying integrated biochemical, biophysical, and spectroscopic approaches to complex metalloenzymes with an emphasis on identifying unique structure-functions relationships and molecular determinates of electron-control. More broadly, the research seeks to uncover basic knowledge that can inform on the design of robust artificial catalysts and new technologies for solar energy conversion to chemical fuels.
Affiliated Research Programs
Photosynthetic Energy Transduction, U.S. Department of Energy's (DOE's) Office of Basic Energy Sciences (BES), Photosynthetic Systems (contributor)
Biological Electron Transfer and Catalysis Energy Frontier Research Center, DOE BES (contributor)
Areas of Expertise
Protein chemistry, biophysical characterization, spectroscopy, and structural biology
Application of integrated spectroscopy (electron paramagnetic resonance [EPR], fourier-transform infrared [FTIR], and X-ray crystallagraphy) for exploring structure-function relationships of redox metalloenzymes
Designated Area Representative of the NREL Spin Resonance Facility, a high-resolution spectroscopic laboratory for multidisciplinary research into electronic, molecular, and atomic structures
Education
Ph.D., Biochemistry, Montana State University, 2010
B.S., Chemistry, Calvin College, 2005
Professional Experience
Scientist IV-Multi Discipline, NREL, 2017–present
Scientist III, Physical Biochemist, NREL, 2014–2017
Postdoctoral Researcher, NREL, 2011–2014
Postdoctoral Research Associate, Montana State University, 2010
Featured Publications
-
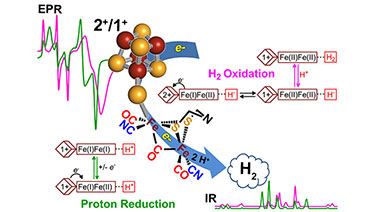
"Investigations on the role of proton-coupled electron transfer in hydrogen activation by [FeFe]-hydrogenase" J. Am. Chem. Soc. (2014)
-
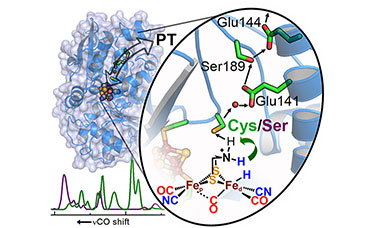
"EPR and FTIR analysis of the mechanism of H2 activation by [FeFe]-hydrogenase HydA1 from Chlamydomonas reinhardtii" J. Am. Chem. Soc. (2013)
-
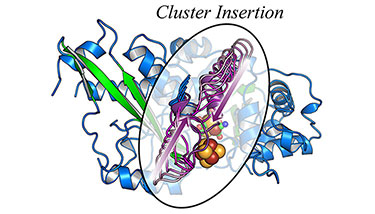
"Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydAΔEFG" Nature (2010)
Additional Publications
-
"The electron bifurcating FixABCX protein complex from Azotobacter vinelandii: Generation of low-potential reducing equivalents for nitrogenase catalysis," Biochemistry (2017)
-
"The reduction potentials of [FeFe]-hydrogenase accessory iron-sulfur clusters provide insights into the energetics of proton reduction catalysis," J. Am Chem. Soc. (2017)
-
"Mechanistic insights into energy conservation by flavin-based electron bifurcation," Nat. Chem. Biol. (2017)
-
"Identification of a Catalytic Iron-Hydride at the H-Cluster of [FeFe]-Hydrogenase," J. Am. Chem. Soc. (2017)
-
"Proton reduction using a hydrogenase-modified nanoporous black silicon photoelectrode," ACS Appl. Mater. Interfaces (2016)
-
"Crystal structure and biochemical characterization of Chlamydomonas FDX2 reveal two residues that, when mutated, partially confer FDX2 the redox potential and catalytic properties of FDX1," Photosynth. Res. (2016)
-
"The effect of a C298D mutation in CaHydA [FeFe]-hydrogenase: Insights into the protein-metal cluster interaction by EPR and FTIR spectroscopic investigation," BBA-Bioenergetics (2016)
-
"[FeFe]-and [NiFe]-hydrogenase diversity, mechanism, and maturation," BBA-Mol. Cell. Res. (2015)
-
"[FeFe]-hydrogenase oxygen inactivation is initiated at the H cluster 2Fe subcluster," J. Am. Chem. Soc. (2015)
-
"Diameter dependent electron transfer kinetics in semiconductor-enzyme complexes," ACS Nano. (2014)
-
"Electron transfer kinetics in CdS nanorod-[FeFe]-hydrogenase complexes and implications for photochemical H2 generation," J. Am. Chem. Soc. (2014)
-
"Hydrogen production by water biophotolysis," in Microbial BioEnergy: Hydrogen Production (2014)
-
"Identification of global ferredoxin interaction networks in Chlamydomonas reinhardtii," J. Biol. Chem. (2013)
-
"Hydrogenases, nitrogenases, anoxia, and H2 production in water-oxidizing phototrophs," (invited book chapter) in Algae for Biofuels and Energy. Springer/Humana Press (2013)
-
"Insights into [FeFe]-hydrogenase structure, mechanism, and maturation," (invited review) Structure (2011)
-
"Small angle x-ray scattering spectroscopy," (invited book chapter) Nitrogen Fixation in Methods Mol Biol. (2011)
-
"Structural basis for carbon dioxide binding by 2-ketopropyl coenzyme m oxidoreductase/carboxylase," FEBS Lett. (2011)
-
"Fad binding by APBE protein from Salmonella enterica: A new class of FAD-binding proteins," J. Bacteriol. (2011)
-
"Activation of HydAΔEFG requires a preformed [4Fe-4S] cluster," Biochemistry (2009)
-
"Hydrogenase cluster biosynthesis: organometallic chemistry nature's way," Dalton Trans. (2009)
-
"New frontiers in hydrogenase structure and biosynthesis," Current Chem. Biol. (2008)
-
"Probing the MgATP-bound conformation of the nitrogenase Fe protein by solution small-angle X-ray scattering," Biochemistry (2007)
View all NREL Publications for David W. Mulder.
Share

